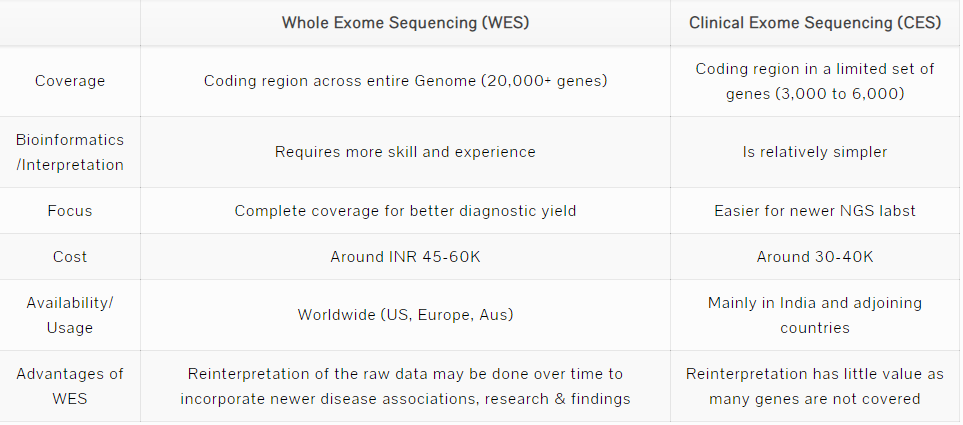
Whole Exome Sequencing (WES) as the name suggests involves sequencing of the Exome or protein coding region of the genome. A WES would cover the coding regions of all the 20,000+ genes. In the absence of a well-defined clinal presentation, WES enables scanning the entire coding region of the genome at a reasonable cost.
Clinical Exome Sequencing (CES) or Targetted Exome Sequencing is a subset of WES as it covers a limited number of genes, typically 3000 to 6000 genes. CES was created by the technology providers (Illumina, ThermoFischer) and NGS labs to make the bioinformatics and interpretation easier. In creating a smaller subset, the idea was to reduce the complexity and costs for the labs and scientists. The subset of genes was chosen based on various studies about the disease associations, prevalence/frequency of mutations in these genes. However, the list of disease-associated genes has been expanding with newer research and publications every week. Hence the variations in the CES gene set of various labs.
While the CES is relatively cheaper and easier for the labs, the underlying assumption is that the non covered genes/regions are not important. Many times, this may lead to missing of a vital mutation.
CES is largely popular in Asian countries and price-sensitive markets. Whereas, the rest of the world is using WES commonly and gradually exploring Whole Genome Sequencing (WGS) which would cover the non-protein coding regions as well.